Breeding livestock has long relied on pedigree information to track ancestry and estimate genetic merit. However, with advancements in genomic technology, breeders now have more precise tools for genetic selection. This article explores the relationship between traditional pedigree-based breeding and genomic breeding and how this also applies to inbreeding.
Pedigree-Based Breeding: Understanding Bloodlines
Pedigree breeding is based on recorded ancestry, tracing lineage through generations. This extends to traditional pedigree inbreeding calculations, which rely on pedigree information in registered populations.
Although the AWA holds the most complete pedigree records for Wagyu, which trace back into the original Japanese registration certificates, these typically truncate 2-4 generations prior to the export of the Foundation animals from Japan in the 1990’s.
We know that there was a significant amount of inbreeding within the Japanese Black population prior to the export of animals and genetics from Japan. Pedigree-based inbreeding calculations cannot account for this.
The AWA recently released Genomic Inbreeding calculations for registered animals with genomic profiles. This analysis showed that using genomic inbreeding calculations, which accurately account for real inbreeding at a DNA level, the average inbreeding within the registered Japanese Black population was around 12%. This is much higher than prior pedigree predicted inbreeding (6%) because it accounts for the prior inbreeding that occurred in Japan before export.
This article will walk through how genomic data can be used to accurately calculate trait performance and actual inbreeding and how genomic data can be used to assist inbreeding management and genetic improvement.
Pedigree breeding and analysis:
This approach assumes that desirable traits are passed down in an averaged and predictable manner, relying on estimated breeding values (EBVs) derived from historical data and performance records and mid-parent (averaged) EBVs.
Key Benefits of Pedigree Breeding:
- Provides a structured method to track family history.
- Helps preserve desirable bloodlines and breed characteristics.
- Aids in estimating genetic merit based on family performance.
However, pedigree breeding has limitations. Because it relies on assumed genetic inheritance rather than measuring the actual DNA that was inherited. Only 50% of each parents DNA is passed on to offspring. Hence, a parent’s full genetic merit is not passed on to its offspring. We can see large variations within full siblings (brothers and sisters) that are not accounted for, potentially leading to unexpected trait expression in progeny.
Likewise for regions of DNA that contain inbreeding, pedigree inbreeding calculations assume an average relatedness and do not account for variation between siblings in inbreeding, which can be equally as large as trait variation between siblings.
Genomic Breeding: A Modern Approach
Genomic selection uses DNA testing to assess the genetic merit of an individual at a molecular level. Instead of relying solely on ancestry, it evaluates thousands of genetic markers across the genome, providing a more accurate prediction of an animal’s genetic potential. It can determine which genetic material is inherited from parents, grandparents and great grandparents.
Advantages of Genomic Breeding:
- Directly measures genetic material rather than inferring it from ancestry
- Identifies animals with superior genetic potential early in life.
- Reduces the uncertainty of breeding decisions by providing precise trait estimates.
- Helps manage inbreeding by assessing actual genetic diversity rather than assumed relationships.
Progeny Relatedness: Parents vs. Grandparents
Each calf inherits 100% of its genetic material from its parents—50% from the cow and 50% from the bull. However, the specific 50% inherited from each parent varies between full siblings, creating genetic diversity even among calves with the same sire and dam.
While full siblings are 100% related by pedigree, meaning they share the same recorded lineage, they are only 50% related by genomic content on average. This difference arises due to Mendelian sampling, a process where the sperm and egg carry a random selection of the parent’s genetic material. As a result, when these gametes combine during fertilization, they create unique genetic combinations in each offspring.
Understanding Genetic Variation Through Mendelian Sampling
The figure below illustrates how chromosome segments are passed from grandparents to parents and then to progeny through recombination during gamete formation. Each parent produces genetically unique sperm or eggs, meaning that no two offspring inherit the exact same DNA combination, even if they share the same parents.
This random genetic recombination results in full siblings having a range of genetic similarity—some may be as low as 35% related while others may be as high as 65% related. The variation is even greater when considering genetic contributions from grandparents, where an individual calf may inherit as little as 20% or as much as 30% from any one grandparent, rather than the expected 25%.

Genomic Inbreeding %
Genomic inbreeding percentage in cattle refers to the degree of genetic relatedness between individuals within a population. It is measured using genomic data and can be used to predict the likelihood that an individual has inherited the same allele from both parents (homozygosity), which determined true inbreeding (loss of genetic variation).
How does it work?
- Genetic Data Collection: To measure genomic inbreeding, DNA samples are taken from the cattle, typically through a tissue sample unit. The genetic information is then analyzed using a genomic test (100K is ideal).
- Marker-based Evaluation: The test looks at specific genetic markers across the animal’s genome. These markers are regions of DNA that vary among individuals. The more markers that are shared between the animal’s parents, the more likely the offspring will inherit the same alleles, leading to homozygosity which is true inbreeding at a DNA level.
- Calculation of Inbreeding Coefficient: Pedigree inbreeding coefficients (F values) are a measure of the probability that two alleles at a given locus in an individual are identical by descent. Genomic inbreeding coefficients are calculated by measuring actual homozygosity across the entire genome and express the proportion of the genome that is homozygous (i.e., the two alleles at a locus are the same) as a percentage.
Genomic inbreeding analysis provides a much more precise and accurate estimate of inbreeding than traditional pedigree-based methods. You can consider the genomic inbreeding is actual inbreeding, whereas pedigree inbreeding is an assumed statistical average.
Implications for Breeding Decisions
In applying genetic tools to assist your breeding decisions, the distinction between pedigree and genomic breeding and inbreeding, has significant implications for efficiency, genetic improvement, and long-term productivity and sustainability.
Pedigree breeding and inbreeding estimates rely on ancestry and expected inheritance patterns but do not account for the variation in actual genetic material passed to offspring. As a result, they may overestimate or underestimate an animal’s genetic potential and inbreeding, having lower relative reliability.
In contrast, genomic breeding and inbreeding tools use precise DNA analysis to identify the specific genetic makeup of each animal, allowing producers to select individuals with superior traits such as carcass traits, growth rates, or genetic conditions and high or low relative inbreeding. This genomic data has higher reliability and increases the accuracy of breeding decisions, accelerating genetic progress within the herd.
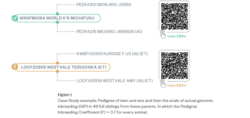

A Case Study: Analysis of inbreeding variation in Full Siblings
In reviewing past articles, we revisited a compelling case from the Australian Wagyu Association (AWA) database, where a breeder repeatedly flushed one of their elite cows to the same sire, resulting in 48 full siblings with genomic data. This unique dataset provides valuable insights into the extent of genetic variation in full siblings and its implications for breeding programs.
The pedigree and genomic inbreeding analysis shown in the figure includes a dataset of49 full siblings from the same dam (LOCFZ0505) and sire (WKSFM0164). This provides a detailed example demonstrating the principle of genetic variation among full siblings. Despite identical parentage, these siblings exhibit significant genomic diversity as demonstrated by the range in genomic inbreeding (measured as homozygosity across the genome).
Of note, is that the predicted pedigree inbreeding coefficient for every single full sibling progeny is 3.7. That is, the pedigree predicted inbreeding is exactly the same for each of the 49 full siblings.
The actual Genomic Inbreeding Coefficient for each full sibling is provided in the figure. The actual genomic inbreeding ranges from a low of 11.8 to a high of 17.7 across the full siblings. This broad range demonstrates that full siblings do not inherit identical genetic material, emphasizing the complexity of genetic recombination and the value of genomics to decipher this variation.
Implications for Breeding Programs
The findings from this study reinforce the importance of accurate genomic selection tools to aid traditional pedigree-based methods. While pedigree-based selection has been a cornerstone of breeding programs, it does not accurately account for the genetic composition of an individual animal. Genomic testing provides a more precise understanding of an animal’s genetic potential, allowing breeders to make more informed selection decisions.
For instance, the variation in marbling performance, growth rates, and carcass yield observed among the full siblings underscores the need for genomic evaluation in breeding programs. By incorporating genomic data, breeders can enhance genetic progress, optimize breeding strategies, and improve overall herd performance.
Using the example of the 49 full siblings and the variation in genomic inbreeding demonstrated here, if a breeder sought to retain certain individuals with low inbreeding as the basis for future breeding animals within their herd, they could significantly reduce the accumulation of inbreeding in future generations by selecting low genomic inbreeding coefficient animals.
This analysis also illustrates the substantial genetic variation that exists even among animals with identical parentage. The variation in genomic inbreeding, carcass performance, and marbling ability highlights the benefits of aiding pedigree-based selection and breeding with accurate genomic tools. As genomic technologies continue to advance, Wagyu breeders and other livestock producers must leverage these tools to maximize genetic gains and improve herd efficiency.
